Cats (Felidae)
The second family described in the database is the Felidae. There are many different species and subspecies in the Felidae family, 12 of them are described in the database. These are the Asian leopard cat, European wild cat, Domestic cat, African wild cat, Cheetah, Cougar, Bobcat, Ocelot, Jaguar, Lion, Leopard and the Tiger. The information in the database consists of the same items as with the Ursidae: the genome sequence, mitochondrial genome sequence, SNP, microsatellites and some literature about genetic studies of that animal. Table 2 in appendix 1 shows a summary of that what is known about the Felidae species, the known genetic information is present in the database. The phylogenetic tree based on the alignment of the whole mitochondrial genome sequence about the Felidae species is shown in figure 10.
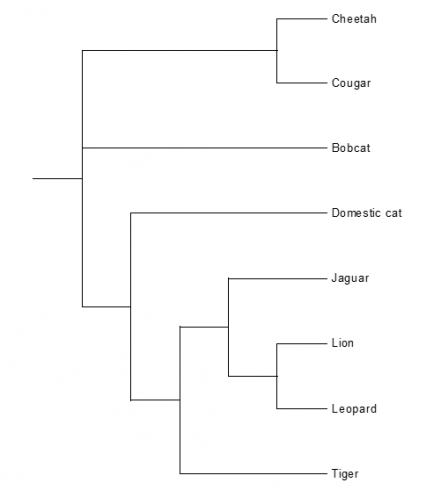
FIGURE 10 Phylogenetic tree based on the whole mitochondrial genome sequence of the Cheetah, Cougar, Bobcat, Domestic cat, Jaguar, Lion, Leopard and the Tiger (species of the Felidae family). This tree is based on the alignment of these animals made in clustalX.
Not all the species are included because the whole mitochondrial genome sequence isn’t known of all the described species of the Felidae. The mitochondrial genome sequence of one animal per species is used to make this phylogenetic tree. The species are: Cheetah, Cougar, Bobcat, Domestic cat, Jaguar, Lion, Leopard and the Tiger. Figure 10 shows which animal species are most related based on the mitochondrial genome sequence. The second phylogenetic tree of the Felidae family is based on the sequence of cytochrome b. For this phylogenetic tree are ten Lions, three Leopards, two Jaguars, ten Tigers, ten Asian leopard cats, nine Domestic cats, two European wild cats, three Cheetahs, two Cougars and three Bobcats. Figure 11 shows which species are most related based on the hypervariable cytochrome b sequence.
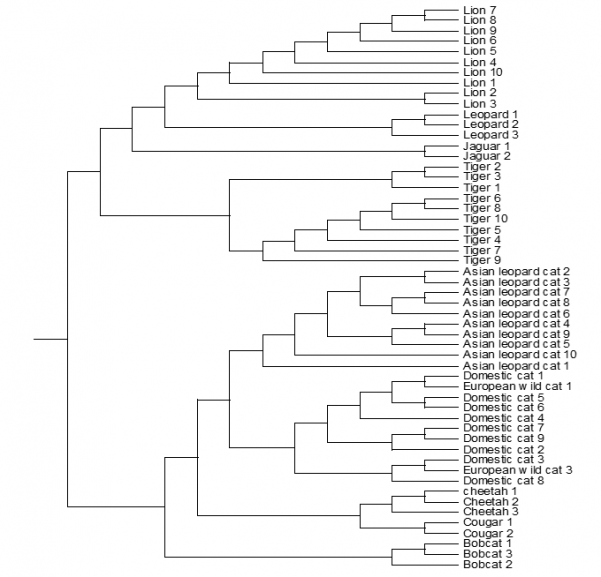
FIGURE 11 Phylogenetic tree based on the cytochrome b sequence of ten Lions, three Leopards, two Jaguars, ten Tigers, ten Asian leopard cats, nine Domestic cats, two European wild cats, three Cheetahs, two Cougars and three Bobcats (species of the Felidae family) This tree is based on the alignment of these animals made in clustalX.
In many species genetic studies have been performed where the genetic relationships have been estimated between animals within a species and animals over species. These analysis need variation detected by sequencing (whole genome, parts of chromosomes , specific genes or gene families, mt DNA). The comparison between animals or species can be displayed in different ways such as in phylogenetic trees. With more genetic/genomic information the phylogenetic tree gets more accurate and will be more reliable.
The two phylogenetic trees in figure 10 and 11 differ from each other (even as the two phylogenetic trees of the Ursidae (bears)). The phylogenetic tree based on the sequence of cytochrome b have more animals per species (figure 11). This phylogenetic tree is more reliable, because there are more animals per species involved. The Lion, Leopard, Jaguar and the Tiger are grouped together and belong to the subfamily Panthera. The other species in the tree are also grouped together and belong to the subfamily Leopardis. Figure 17 gives the conclusion of the cat phylogenetic tree made by Collier & O'Brien.
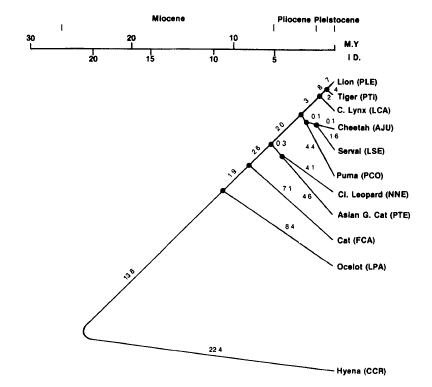
FIGURE 17 Phylogenetic tree using the Fitch-Mar-goliash algorithm(1 967) and the FITCH program of Felsenstei. Based on the immunologic distance (Collier & O'Brien, (May, 1985)).
This tree is based on the immunologic distance. Collier & O'Brien used only one animal per species. This phylogenetic tree is first of all different from the tree in figure 17 because the trees contain some different spieces. The phylogenetic trees also differ in the information were they are based on. Figure 17 is based on the immunologic distance and only one animal per species. The phylogenetic tree in figure 11 is based on the sequnce of cytochrome b and is based on multiple animals per spieces. This can be the reason why the Cheetah and the Leopard are placed in a different way in the phylogenetic tree (difference in figure 11 and 17). There are more researches done about the part of the big cats (panthera) in the phylogenetic tree.
The conclusion of Davis, Li, and Murphy is shown in figure 18.
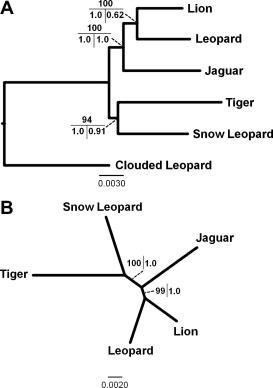
FIGURE 18 Maximum likelihood (ML) tree based on analysis of the complete supermatrix. (A) Rooted with clouded leopard as outgroup. 1000 ML bootstrap replicate percentages depicted on the top, Bayesian posterior probabilities (BPP) on the bottom left, and BEST posterior probabilities on the bottom right. (B) Unrooted topology with ML bootstrap percentages on the left and BPP on the right (Davis, Li, & Murphy, 2010).
This phylogenetic tree consists only the panthera species: Lion, Leopard, Jaguar, Tiger, Snow leopard and the Clouded Leopard (figure 18). Only the Snow Leopard and the Couded Leopard are not included in the phylogenetic tree in figure 11. The species that are the same in both phylogenetic trees are also placed in the same order in the phylogenetic trees. The phylogenetic tree of Davis, Li and Murphy (Figure 18) is based on more genetic information. They used parts of the autosomal genome sequence, Y-chromosomal sequence and parts of the mitochondrial genome sequence. This is more information than is used for figure 11, but they are the same. This proves that this part of the cat phylogenic tree is in this way. Masuda et al. concentrated solely on the other part of cat phylogeny. They concentrated on the Domestic cat lineage and the ocelot lineage, but also included some species of the pantherine lineage.
They based their phylogenetic tree also on the sequence of cytochrome b (Figure 19).
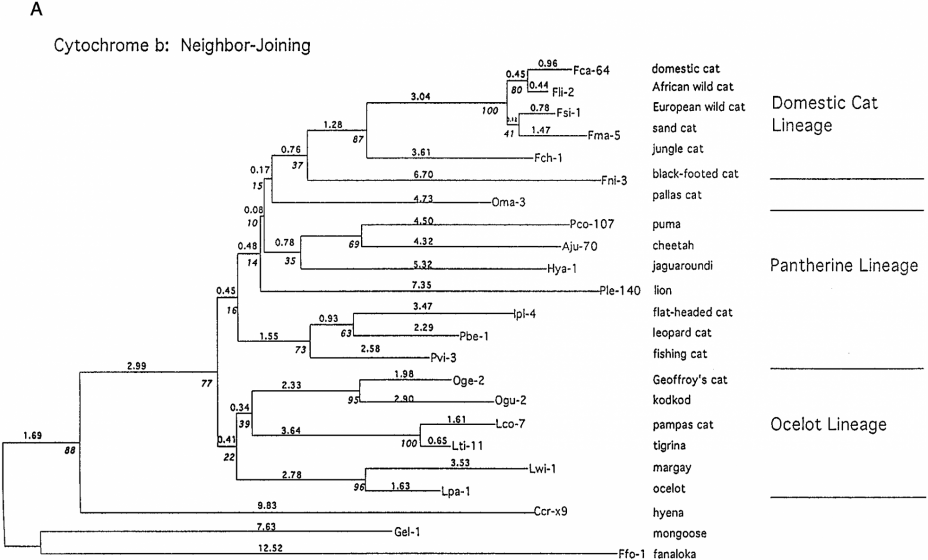
FIGURE 19 Phylogenetic reconstruction using minimum evolution estimated by neighbor-joining method of cytochrome b sequences. Trees were derived using Kimura genetic distance estimates (Masuda, Lopez, Slattery, Yugki, & O’Brien, 1996).
The species that are the same as in figure 11 are organised almost the same in the phylogenetic tree. Only the Leopard cat is in a different order, but it is not clear if in the research of Masuda et al. also the Asian leopard cat is used or another leopard cat species.