The first family of the order Carnivora that is described in the database is the Ursidae. The Ursidae family consist of eight species. These are the Polar bear (Ursus martinimus), Brown bear (Ursus arctos), American black bear (Ursus americanus), Asiatic black bear (Ursus thibetanus), Sun bear (Ursus malayanus), Sloth bear (Ursus ursinus), Spectacled bear (Tremarctos ornatus) and the Giant panda (Ailuropoda melanoleuca). The information in the database consists of the genome sequence, mitochondrial genome sequence, SNP, microsatellites and some literature about that animal. Table 1 in Appendix 1 shows a summary of what is known about the Ursidae species, the known genetic information is present in the database. The alignment made in clustalX about the species of the Ursidae is shown in figure 7. This is only a part of the alignment. This alignment is from the whole mitochondrial genome sequence with only one animal of every species of the Ursidae families. Figure 8 represent the phylogenetic tree made in TreeView on the basis of the alignment shown in figure 7.
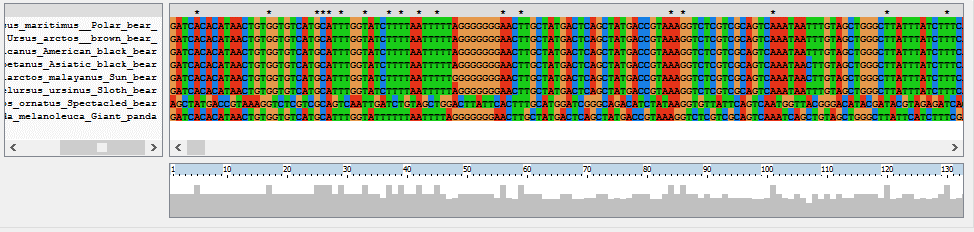
FIGURE 7 A part of the alignment from the whole mitochondrial genome sequence of the speceies out of the Ursidae familie made in ClustalX
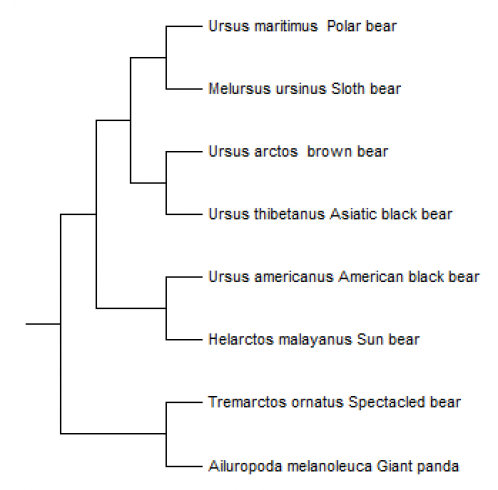
FIGURE 8 Phylogenetic tree made in tree view on the basis of the alignment in clustalX of the mitochondrial genome sequence of the species out of the Ursidae family. The mitochondrial genome sequence of one animal per species is used for this tree.
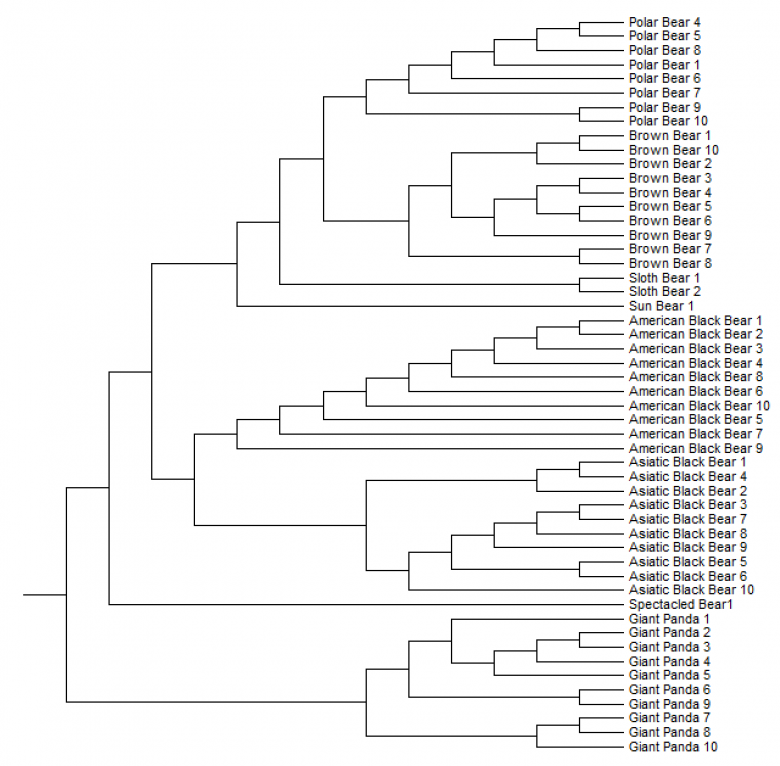
FIGURE 9 Phylogenetic tree of the cytochrome b sequence of multiple animals per species (eight Polar bears, ten Brown bears, two Sloth bears, one Sun bear, ten American black bears, ten Asiatic black bears, one Spectacled bear and ten Giant pandas). This tree is based on the alignment of these animals made in clustalX.
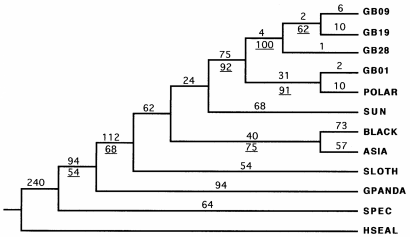
FIGURE 14 Single most parsimonious phylogenetic tree inferred by weighted parsimony methods based on the combined cytochrome b/ tRNAPro and tRNAThr genes of eight species of Ursidae and using harbor seal as outgroup. Labels are as follows: H. SEAL= harbor seal, GPANDA= giant panda, SPEC = spectacled bear, SUN =sun bear, SLOTH= sloth bear, ASIA=Asiatic black bear, BLACK= American black bear, BROWN = brown bear, and POLAR = polar bear. Lineages of brown bear are as follows: GB01 =islands of southern Alaska, GB09 = coast of south-eastern Alaska, GB19= western Alaska, GB28 =Turkey (Talbot & Shields, 1996).
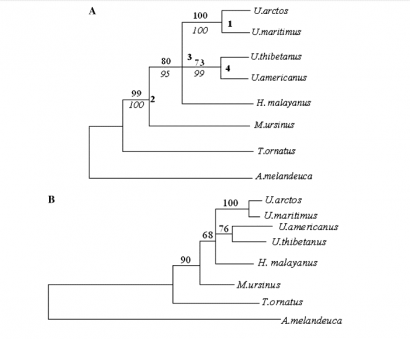
FIGURE 15 Phylogenetic hypotheses based on analyses of concatenated nuclear and mt sequences. Cladogram A (A) shows maximum parsimony (MP) tree. Phylogram B (B) shows maximum likelihood (ML) tree (Yu, Li, Ryder, & Zhanga, 2004)
The second phylogenetic tree of the cytochrome b sequence of multiple animals per species of the Ursidae is shown in figure 9. For the alignment the cytochrome b sequence of eight Polar bears, ten Brown bears, two Sloth bears, one Sun bear, ten American black bears, ten Asiatic black bears, one Spectacled bear and ten Giant pandas were used. The animals of the same species are clustered together in the phylogenetic tree.
In many species genetic studies have been performed where the genetic relationships have been estimated between animals within a species and animals over species. These analysis need variation detected by sequencing (whole genome, parts of chromosomes , specific genes or gene families, mt DNA). The comparison between animals or species can be displayed in different ways such as in phylogenetic trees. With more genetic/genomic information the phylogenetic tree gets more accurate and will be more reliable.
The two different phylogenetic trees made (figure 8 and 9), differ in a particular way. In the first one (figure 8) the Polar bear and the Sloth bear are most related, the Brown bear and the Asiatic black bear are most related, the American black bear and the Sun bear are most related and the spectacled bear and the Giant polar are most related to each other. The second phylogenetic tree (figure 9) the Polar bear and the Brown bear are most related. This two together are than most related to the Sloth bear. These three species together are than most related to the Sun bear. The American and the Asiatic black bear are most related together, and these species all together are most related to the Spectacled bear. At last they are related to the Giant panda. The last one is more reliable, because this phylogenetic tree include more animals per species. There is research with other interpretations of the phylogenetic tree of the Ursidae. Talbot and Shields (1996) concluded that the most uncertain relationships among phylogenies of Ursidae are: (1) whether the Giant panda (Ailuropoda melanoleuca) should be included among the Ursidae; (2) the location of the spectacled bear (Tremarctos ornata) in relation to the ursines; (3) the hierarchical order of the divergence among the six ursine bears; and (4) whether the Brown bear (Ursus arctos) is paraphyletic with respect to the Polar bear (Ursus maritimus). The conclusion of Talbot and Shields (1996) is shown in figure 14. They used the sequence of the cytochrome b, tRNAPro and tRNAThr genes. The difference between the phylogenetic tree based only on the cytochrome b sequence (figure 9) and the phylogenetic tree of Talbot and Shields (1996) (figure 14) is that the Spectacled bear and the Sloth bear are on different places in the tree.
The reason of this can be that in figure 9 only the sequence of one Spectacled bear and two of the Sloth bear are used. Figure 14 is based on the sequence of three different bears of each. With more genetic information is the phylogenetic tree more reliable.
Yu et al. (2004) came to another conclusion of the phylogenetic tree (figure 15).
This analyses is based on the nuclear and the mitochondrial DNA sequences. The Sloth bear (M. Ursinus) and the Sun bear (H. malayanus) are on a different place in the tree in this research than in the phylogenetic tree made of sequence of cytochrome b (figure 9). The mitochondrial sequence described only the mother line of the DNA, Yu et al. (2004) combined the autosomal and the mitochondrial sequences. This can explain why the phylogenetic trees are different from each other. Another conclusion of the phylogenetic tree can be found in figure 16. This is a phylogenetic tree based on 14 nuclear genes, and represents the conclusion of Page et al. (2008).
This tree should be different from the phylogenetic tree in figure 15, because in figure 15 only the mother line is described in the phylogenetic tree. There are some differences between the trees. The Asiatic black bear, Sloth bear and the Sun bear are placed in another orientation in the different trees. The first point of uncertain relationships among phylogenies of Ursidae was if the Giant panda should be included in the Ursidae. The Giant panda is included in all the phylogenetic trees showed here. The Giant panda belongs to the Ursidae and not in the Procyonids or in another separated group (Sarich, 1973).
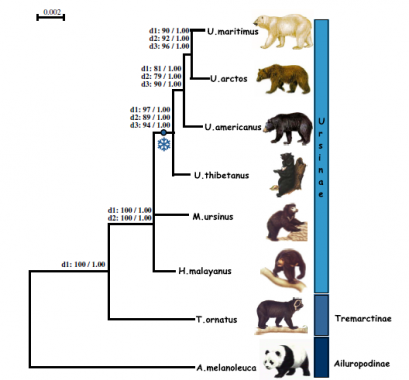
FIGURE 16 Phylogenetic tree of the Ursidae family based on the analyses of 14 combined nuclear genes and reconstructed following Bayesian methods (Page, Calvignac, Catherine, & Paris, 2008).